Sequencing - Nemabiome.ca
Sequencing Protocols
Download a pdf of all the protocols for printing here
Amplication of ITS-2 region
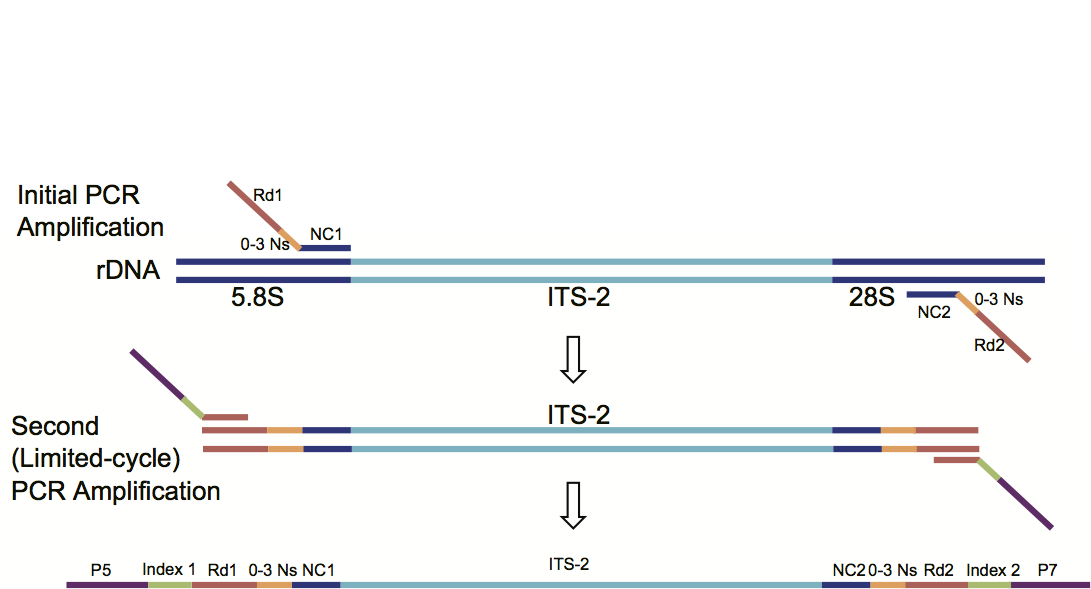
Reagents required
- Kapa HiFi Hotstart PCR kit with dNTPs (Kapa Biosystems, KK2502)
- 0.2ml 96-well plates, clear, half-skirted (VWR, 83007-374)
- Microseal “A” film (Biorad, MSA-5001)
- 1.5 ml tubes
- 8-channel pipetter
- filter aerosol tips
- mol grade water
- thermocycler
| Name | Sequence |
|---|---|
| NC1_with_Illumina_Adapter_(0N) | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGACGTCTGGTTCAGGGTTGTT |
| NC1_with_Illumina_Adapter_(1N) | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGNACGTCTGGTTCAGGGTTGTT |
| NC1_with_Illumina_Adapter_(2N) | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGNNACGTCTGGTTCAGGGTTGTT |
| NC1_with_Illumina_Adapter_(3N) | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGNNNACGTCTGGTTCAGGGTTGTT |
| Name | Sequence |
|---|---|
| NC2_with_Illumina_Adapter_(0N) | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTTAGTTTCTTTTCCTCCGCT |
| NC2_with_Illumina_Adapter_(1N) | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGNTTAGTTTCTTTTCCTCCGCT |
| NC2_with_Illumina_Adapter_(2N) | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGNNTTAGTTTCTTTTCCTCCGCT |
| NC2_with_Illumina_Adapter_(3N) | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGNNNTTAGTTTCTTTTCCTCCGCT |
Protocol
- UV treat plasticware and mol grade water for 15 min prior to PCR set-up
- Combine 2.5 μl of each of the forward primers at stock concentration (100 nM) to 90 μl of mol grade water to achieve forward primer mix at 10nM working concentration. Do the same for the reverse primers.
- On ice make-up Mastermix using forward and reverse primer mixes and the recipe below.
- Aliquot mastermix (21 μl) to each well of 96-well plate
- Add 4 μl template (lysate dilution) to wells using multi-channel pipetter
- Place microseal “A” film over completed plate and place on thermocycler (with pre-heated lid) and run the program below.
| volume (μl) | Reagent |
|---|---|
| 5 | KAPA HiFi Buffer (x5) |
| 0.75 | NC1 primer mix (10mM) |
| 0.75 | NC2 primer mix (10mM) |
| 0.75 | dNTPs (10mM) |
| 0.5 | KAPA HiFi Polymerase (0.5U) |
| 13.25 | mol grade water |
| 4 | 1:10 dilution of lysate |
Cycling conditions
- 95 for 2 min
- 98 for 20 sec
- 62 for 15 sec
- 72 for 15 sec
- Repeat steps 2-4 25 times
- 72 for 2 min
Cycles kept low to limit potential bias, but can be increased to 30-35 cycles for samples with low DNA. If amplicons are not to be cleaned immediately store at -20. It is not advisable though to delay the library preparation process as PCR products will degrade even at -20 if stored for longer than a couple of days
Magnetic bead purification
Reagents required
- AMPure XP beads (Beckman Coulter, A63881) - Aliquot and store at 4∘C
- 96-well Magnetic-Ring stand (Applied biosystems, AM10050)
- 0.8 ml 96-well storage plates (Thermo-Scientific, AB-0859)
- 0.2 ml 96-well plate, clear, half-skirted (VWR, 83007-374)
- 8-channel pipetters
- Reagent reservoirs
- absolute ethanol (mol grade)
- mol. grade water
Filter aerosol pipette tips and 8-channel pipetter used for majority of steps in this protocol. Also all quantities quoted are for clean-up of a full plate (96 samples)
Protocol
- Bring an aliquot of AMPure beads to room temp
- Prepare fresh 80% ethanol from mol grade absolute ethanol (50
ml)
- Centrifuge amplification plate to collect condensation (300 G for 2
min)
- Remove seal from plate in biosafety cabinet (if possible)
- Transfer the 25 μl reactions
into 0.8 ml 96-well storage plates using multi-channel pipettor
- Voretx beads for 30 sec to ensure even disperal before placing in
reagent reservoir
- Transfer 25 μl of beads to
each well of the storage plate and gently pipet up and down 10 times to
ensure the beads are well-mixed with the PCR sample
- Incubate at room temp for 5 min
- Place plate on magnetic stand for 2 min (or until supernatent has cleared)
- Once clear use multi-channel pipettor to remove and discard
supernatent from the wells
- With the plate still on the magnetic stand, wash beads with 80%
ethanol by adding 200 μl to each
well (using a multi-channel pipetter and a reagent reservoir). Do not
resuspend the beads at this point
- Incubate for 30 sec (or until clear) and then remove and discard
supernatent
- Wash again with another 200 μl of 80% ethanol. Incubate for 30 sec
and then as before remove and discard supernatant.
- Use a small volume single-channel pipettor (P20, with fine tips) to
remove excess ethanol from each well
- Allow beads to air-dry for 15 min
- Remove plate from magnetic stand
- Add 40 μl of mol grade water
to each well with multi-channel pipettor and gently pipet up and down 10
times to re-suspend the beads
- Incubate at room temp for 2 min
- Place the plate back on the magnetic stand for 2 min (or until
cleared)
- Transfer approx 34 μl of the
supernatent to a new 0.2 ml 96-well plate without touching the
beads
- A microseal “A” is placed over cleaned-up samples to allow temporary storage at −20∘C
Addition of Illumina barcodes
Reagents required:
- Kapa HiFi Hotstart PCR kit with dNTPs (Kapa Biosystems, KK2502)
- 0.2 ml 96-well plates, clear, half-skirted (VWR, 83007-374)
- Microseal “A” film (Biorad, MSA-5001)
- 1.5 ml tubes
- 8-channel pipettor
- filter aerosol tips
- mol grade water
- thermocycler
| Primer name | Sequence of primer | Row in 96-well plate |
|---|---|---|
| N501_i5 | AATGATACGGCGACCACCGAGATCTACACTAGATCGCTCGTCGGCAGCGTC | A |
| N502_i5 | AATGATACGGCGACCACCGAGATCTACACCTCTCTATTCGTCGGCAGCGTC | B |
| N503_i5 | AATGATACGGCGACCACCGAGATCTACACTATCCTCTTCGTCGGCAGCGTC | C |
| N504_i5 | AATGATACGGCGACCACCGAGATCTACACAGAGTAGATCGTCGGCAGCGTC | D |
| N505_i5 | AATGATACGGCGACCACCGAGATCTACACGTAAGGAGTCGTCGGCAGCGTC | E |
| N506_i5 | AATGATACGGCGACCACCGAGATCTACACACTGCATATCGTCGGCAGCGTC | F |
| N507_i5 | AATGATACGGCGACCACCGAGATCTACACAAGGAGTATCGTCGGCAGCGTC | G |
| N508_i5 | AATGATACGGCGACCACCGAGATCTACACCTAAGCCTTCGTCGGCAGCGTC | H |
| Primer name | Sequence of primer | Column in 96-well plate |
|---|---|---|
| N701_i7 | CAAGCAGAAGACGGCATACGAGATTCGCCTTAGTCTCGTGGGCTCGG | 1 |
| N702_i7 | CAAGCAGAAGACGGCATACGAGATCTAGTACGGTCTCGTGGGCTCGG | 2 |
| N703_i7 | CAAGCAGAAGACGGCATACGAGATTTCTGCCTGTCTCGTGGGCTCGG | 3 |
| N704_i7 | CAAGCAGAAGACGGCATACGAGATGCTCAGGAGTCTCGTGGGCTCGG | 4 |
| N705_i7 | CAAGCAGAAGACGGCATACGAGATAGGAGTCCGTCTCGTGGGCTCGG | 5 |
| N706_i7 | CAAGCAGAAGACGGCATACGAGATCATGCCTAGTCTCGTGGGCTCGG | 6 |
| N707_i7 | CAAGCAGAAGACGGCATACGAGATGTAGAGAGGTCTCGTGGGCTCGG | 7 |
| N708_i7 | CAAGCAGAAGACGGCATACGAGATCCTCTCTGGTCTCGTGGGCTCGG | 8 |
| N709_i7 | CAAGCAGAAGACGGCATACGAGATAGCGTAGCGTCTCGTGGGCTCGG | 9 |
| N710_i7 | CAAGCAGAAGACGGCATACGAGATCAGCCTCGGTCTCGTGGGCTCGG | 10 |
| N711_i7 | CAAGCAGAAGACGGCATACGAGATTGCCTCTTGTCTCGTGGGCTCGG | 11 |
| N712_i7 | CAAGCAGAAGACGGCATACGAGATTCCTCTACGTCTCGTGGGCTCGG | 12 |
Protocol
- UV pasticware and mol grade water for 15 min prior to PCR set-up
- On ice make-up Mastermix of water, buffer and dNTPs only (no polymerase or primers yet).
- Aliquot mastermix (19 μl) to each well of 96-well plate
- Add 1.25 μl forward primer to each well of the designated row (A-H, see table above).
- Add 1.25 μl reverse primer to each well of the designated column (1-12, see table above).
- Add 0.5 μl polymerase to each well
- Add 3 μl template (amplicons from the 1st round of PCR) to wells using multi-channel pipettor
- Place microseal “A” film over completed plate and place on thermocycler (with pre-heated lid) and run the program below.
- The resulting PCR products are cleaned-up using the magnetic bead purification protocol similiar to the 1st round of PCR (see section 3.2 above).
| Volume (μl) | Reagent |
|---|---|
| 5 | KAPA HiFi Buffer (x5) |
| 1.25 | Forward primer, N501-508 (10mM) |
| 1.25 | Reverse primer, N701-712 (10mM) |
| 0.75 | dNTPs (10mM) |
| 0.5 | KAPA HiFi Polymerase (0.5U) |
| 13.25 | mol grade water |
| 3 | 1st round PCR product |
Cycling conditions
- 98 for 45 sec
- 98 for 20 sec
- 63 for 20 sec
- 72 for 2 min
- Repeat steps 2-4 7 times
Cycles kept low to limit potential bias
Individual sample quantification and pooling
- Nanodrop quantification of cleaned-up products resulting from second round of PCR
- Make normalized library by combining samples into a single tube to ensure all samples are present in equal concentrations (typically combine 50 ng per sample).This should ensure uniform coverage of reads for all samples in the library.
- Nanodrop library and dilute with mol grade water to achieve approx 5 ng/μl
Reagents required:
- Illumina library Quantification Kit, Universal qPCR Mix (Kapa Biosystems, KK4824)
- qPCR machine
- mol. grade water
- 10 mM Tris-Hcl, pH 8 with 0.05% Tween-20
- Multiplate PCR plate, low profile, clear, unskirted, (Biorad, MLL9601)
- Microseal “B” seal (Biorad, MSB1001)
When setting-up qPCR reactions always take care when pipetting small volumes and use filter aerosol pipette tips. Work on ice and use foil to prevent UV degradation of sybr green in master mix
Protocol
- Dilute library (1:1000) with 10 mM Tris-Hcl, pH 8 with 0.05% Tween-20 in triplicate
- Using these 1:1000 dilutions set-up 1:2 serial dilutions to achieve 1:2000, 1:4000 and 1:8000 (keep in fridge until needed)
- Before using the Illumina library quantification kit for the first time add 1ml of primer premix (x10) to the 5ml bottle of Sybr fast qPCR Master mix (x2) and mix well
- UV all plasticware and water for 15 min
- Prepare qPCR plate by adding:
- 12 μl Sybr fast qPCR
mastermix (with primers added)
- 4 μl mol grade water
- and either 4 μl diluted library, one of supplied standards (Standard 1-6) or another 4 μl of mol grade water to act as a negative control
- 12 μl Sybr fast qPCR
mastermix (with primers added)
- Place Microseal “B” seal over reactions
- Run on qPCR machine with the following cycling conditions: −95∘C for 5 min then 35 cycles of 95∘C for 30 sec and 60∘C for 45 sec
- Look at data to confirm 90-100% reaction efficiency for samples and for standards
- Calculate library quantification using absolute quantification against the 425 bp DNA standard
Reagents required
- mol. grade water
- 1.5 ml eppendorf tubes
- Ice
- 5 M NaOH
- Vortex
- Bench-top centrifuge
- 10 mM Tris-Cl, pH 8.5 with 0.1% Tween-20
- Hybridization buffer (HT1, thawed and kept on ice)
- PhiX control v3 (Illumina, FC-110-3001)
Protocol
- Dilute library to 4 nM with water
- Prepare 1ml fresh 0.2 M NaOH from 5 M NaOH stock solution
- Denature 4 nM library:
- add 5 μl 4 nM library with 5 μl 0.2 M NaOH in new 1.5 ml tube
- vortex briefly and centrifuge @ 300 g for 1 min
- incubate at room temp. for 5 min
- add 990 μl hybridization buffer (chilled) to produce 20 pm library
- place on ice
- Dilute 20 pm library to 15 pmol:
- add 900 μl 2 0pmol library to new 1.5 ml tube
- add 300 μl hybridization buffer (chilled)
- invert several times and place on ice
- Prepare 4 nM PhiX library:
- add 2 μl 10 nM PhiX library to new 1.5 ml tube
- add 3 μl 10 mM Tris-Cl, pH 8.5 with 0.1% Tween-20
- Denature 4 nM PhiX library:
- add 5 μl 4 nM PhiX library with 5 μl 0.2 M NaOH to new 1. 5ml tube
- vortex briefly and centrifuge @ 300 g for 1 min
- incubate at room temp. for 5 min
- add 990 μl hybridization buffer (chilled) to produce 20 pM PhiX library
- place on ice
This denatured 20 pM PhiX library can be stored at −20∘C for up to 3 weeks
- Dilute 20 pM PhiX library to 15 pM:
- add 225 μl 20 pM Phix Library to new 1.5 ml tube
- add 75 μl hybridization buffer (chilled)
- invert several times and place on ice
- Combine 15 pM libraries (80% sample library: 20% PhiX library):
- add 800 μl 15 pM sample library to new 1.5 ml tube
- add 200 μl 15pM PhiX library
- invert several times and place on ice until ready for loading into cartridge
Reagents required
- Illumina Miseq v2 Reagent Kit, 500 cycles (MS-102-2003, contains 2
boxes)
Box 1, contains Reagent cartridge and hybridization buffer (HT1)- store at −20∘C
Box 2, contains Flow cell and Incorporation buffer (RP2) - store at 4∘C - water bath
- tweezers
- mol grade water in squeezy bottle
- kim wipes
- ethanol wipes
Protocol
- Thaw cartridge in water bath at room temp. for 1 hour
- Invert cartridge 10 times to mix reagents and bang down on bench to eliminate any visible air bubbles.
- Wipe foil covering of position 17 before piercing with a 1 ml pipette tip.
- Load 600 μl of the “combined library” into the cartridge (position 17)
- Start software and delete previous run to ensure >25% memory space on machine
- Prepare and load flow cell
- use tweezers to remove flow cell from salt solution.
- wash-off salt with mol. grade water in a squeeze bottle, focusing on the nooks and crannies
- dry thoroughly with kim wipe
- use ethanol wipe to gently remove dust and grease stains from the flow cell
- remove old flow cell from machine and place new one into position
- Load incorporation buffer and empty waste bottle into position and lower sippers
- Load cartridge and csv sample sheet file
